Fundamental Studies of Rheumatoid Arthritis Genetics & Genomics
RA Genetics 101: Glass half-full or half-empty? Part 3
This is Part 3 of a guest series on RA genetics by Robert West, Jr, PhD.
Part 1 provided the introduction to RA genetics and Part 2 described the direct to consumer (DTC) genetics of RA. Here, I describe a dissection of the complexity of the heritable component of RA risk, vis a vis that provided by 23andMe that was discussed in Part 2, estimated to be between 50% and 60% of the total RA risk. Kelly alluded to the topic of genetic risk briefly in a blog post back in October. Environmental contributions which would comprise the remaining 40% to 50% of the total risk of RA have been discussed at length elsewhere, including on RA Warrior.
As regular readers of the RA Warrior blog are aware, RA is an ill-defined chronic condition affecting at least a million and a half Americans, the majority being women. In fact, female gender is the single, largest genetic risk factor for RA. Unfortunately, a scientific explanation for this association still is unavailable. Notwithstanding this shortcoming, with the completion of the human genome project, and the construction of the Human Hapmap, it has been possible to identify other risk factors, specific gene regions (“loci”) (expandable map) if not particular genes themselves, linked to risk of developing common, polygenic diseases such as RA. Twin studies originally showed that RA was heritable and persists in families, and moreover, genetically overlaps to certain extents with other specific autoimmune diseases. Heritability established by classical genetic methods is what provides an indication to proceed with modern genomic approaches to studying RA.
One prominent genomic methodology for probing the association between common human genetic variation and particular human diseases is termed Genome-wide Association Study (GWAS). Most common, chronic human disease conditions have now been analyzed with this method, and a number of candidate genes or loci linked to each condition identified (ibid). Unsurprisingly, instances of overlap of gene association for more than one disease are often observed. This will be the topic of Part 5 of this series, where I will discuss the issue of genetic overlap among autoimmune diseases and its potential impact on clinical diagnosis and treatment of RA. This becomes the nuts and bolts for a new era of personalized medicine, which is the thrust of my present occupation.
In the case of RA, multiple GWAS have been performed within the past decade, and corresponding common risk susceptibility genes/loci identified. Figure 1 at the previous link shows an example of more than 30 common variants in genes or loci that impact risk of RA have been discovered in this way. Of this total, approximately 9 genetic loci, often regarded as among the key risk factors for RA susceptibility, are listed in Table 1 below.
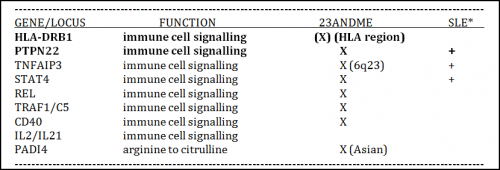
Eight of these nine are relevant to those of European descent, and significantly, all 8 are thought to be involved in some aspect of immune cell signaling. This is not a nominal finding, since genetic association studies often identify variants in loci devoid of recognizable genes, or at best, the inferred biological function of a linked gene may have no obvious connection to the disease being studied. This provides a special advantage to RA patients who are desperate for progress towards new therapies.
The ninth gene listed in Table 1, PADI4, is associated with a common genetic variant linked to RA susceptibility only in a Japanese population. Interestingly, however, PADI4 encodes an enzyme responsible for conversion of the amino acid arginine to citrulline, an activity thought to be specific to and important for protein modification related to proper disposal of dying cells. The latter process is suspected of being responsible for generating “non-self” antigens that invoke an autoimmune response in individuals who have the ACPA+ (anti-citrullinated peptide antibodies) subtype of RA (demonstrated by a positive anti-CCP test). I will elaborate further on this issue in Part 4 of this series where I discuss subtypes of RA.
It is noteworthy that 23andMe presently tests 8 of the 9 common genetic variants listed in Table 1. Of them, a common variant at HLA-DRB1 is associated with the largest heritability effect on RA, particularly ACPA+ RA. This comes as no real surprise, however, since it’s long been known that the human major histocompatibility locus (MHC), where HLA-DRB1 is located, plays a major role in immune cell function, including autoimmunity. A common variant of next highest impact is present in the gene PTPN22, known to encode a protein tyrosine phosphatase enzyme important to T cell function (see also Science Direct). Combined, common genetic variants associated with HLA-DRB1 and PTPN22 account for the vast majority of the known genetic risk of susceptibility to RA (see also Oxford Journal of Rheumatology and Fig.1 here). Indeed, HLA-DRB1 itself accounts for 30 to 40% of the overall genetic susceptibility to RA (see also Genetics of Rheumatoid Arthritis: Time for a Change and Genetics of ACPA positive RA: the beginning of the end?). Meanwhile, common variants for all of the other ~30 genes/loci impacting risk of RA, when combined, account for only ~ 5% of the total genetic risk.
While 23andMe presently tests for a common variant in the “HLA region” that appears to be significantly linked to RA risk, it presently does not specifically test the common variant located at HLA-DRB1, at least with the older V2 chip (see Part 2), though it may test common variants nearer by this locus with the newer V3 chip. This being the single most significant risk location for RA, and HLA-DRB1 or else common risk variants near other genes in the MHC significantly impacting most autoimmune diseases, I would anticipate that 23andMe will attempt to offer direct HLA-DRB1 testing as part of their future genotyping service. Interestingly, another DTC genetic testing company, deCODEme, does test for a common variant more directly associated with HLA-DRB1. On this note, details regarding which of the common genetic variants affecting RA risk presently tested by each of three major DTC genetic testing companies is provided in Table 2 below, obtained from DIYgenomics (gratis Melanie Swan). This table, unlike the simpler one above, includes each specific SNP variant (rs number; described in Part 2) associated with RA risk, as well as several additional details such as chromosomal location and major vs minor allele at the respective gene or locus.

Note: Another resource for determining which diseases are tested by these 3 genetic testing companies is the Gene-Disease Associations table provided by the Genomic Medicine Initiative from the Department of Pathology at Beth Israel Deaconess Medical Center.
Potential contribution of genetics to Rheumatoid Arthritis risk
I will not discuss any of the other common variants in genes/loci shown to be associated with risk of developing RA since their overall impact on genetic risk of RA is quite small (odds ration less than 1.5 each). More detailed information on each of them can be obtained from the specific references provided above, as well as links attached to Table 2.
It is indeed important to mention, however, that additional information on each individual common risk variant, aka single nucleotide polymorphism (SNP), can be obtained at a valuable online resource called SNPedia. This is achieved simply by copy/pasting the respective rs number into the search window provided at the SNPedia website. Alternatively, typing “rheumatoid arthritis” into the search window in SNPedia will bring up a page that includes most of the published common variants found to be associated with this disease. One of the variants listed therein, rs6457617, is the one tested by 23andMe located in the HLA region (see also large genome-wide association study in Nature. Notably, in the homozygous defective state (TT) this variant increases risk of RA by 5.2x.
From the above discussion, it would seem that testing of either HLA-DRB1 (rs660895) by deCODEme or HLA region (rs6457617) by 23andMe would provide a significant indication of potential susceptibility of RA from that locus alone. Yet, bear in mind that common genetic variants represent risk factors that of themselves, and often even when present in combination, are not solely determinative of disease onset. Other risk alleles, scattered across the genome, some of which may have a protective capacity, are likely involved, and disease risk is also impacted by environmental factors (40 to 50% of the total risk of RA). This issue will be discussed in detail in Parts 4 and 5, where the potential predictive capability of RA genetics, particularly for pre-determinative diagnostic purposes, is described. Moreover, the above discussion has been confined to solely common risk variants. Potentially equally important contributions might be made by rare genetic variants which have yet to be uncovered and might be associated with any one of the ~ 30 known RA susceptibility genes or even elsewhere in the genome. Such rare variants are expected to be revealed in the near future, when whole genome sequencing becomes routinely more clinically available.
Practical uses for genetic study of Rheumatoid Arthritis?
So, of what practical use would the various genetic and genomic results presented above be to people suffering with RA? My answer can be divided into 3 sentiments: The Good, The Bad, and The Beyond Pathetic.
The Good: Genetic variants have now been identified that can be used as fish hooks to identify genes, protein, and molecular pathways for the purpose of designing specific drugs. As Eric Lander said recently, the primary goal of scientific studies such as those described above is “to understand the biological pathways underlying disease, because that’s what’s fundamental to treating disease” while “a secondary goal is to be able to provide individual risk prediction for disease; this goal is maybe only partially feasible and is, in my opinion, less important.” This is where we start to enter a glass half-full vs. half-empty argument. Identifying biological targets for drug (or gene) therapy is categorically extremely important if not essential to make progress towards treating RA. Moreover, given genetic overlap among various autoimmune diseases (described in Part 5), progress on understanding and treating RA will have a spillover effect on other chronic, disabling diseases.
Even though genetic data like those provided DTC by 23andMe and other genetic testing companies are presently limited in their predictive value, this is a very new technology and we may expect refinements that ultimately improve their predictability. Improved predictive power might be expected to be achieved, for example, once large numbers of patients become enrolled in electronic health records (EHRs) available for mining of both genomic (e.g. SNP variant) and clinical information (see, for example, Kaiser’s Massive Genetic Database). Meanwhile, knowledge of personal genetic information linked to common disease contributes to the empowerment of the individual who secures such information, which is no small matter in an age of increasing patient advocacy.
The Bad: Unfortunately, it can take years to develop a specific and effective drug once a biological target has been identified. This will leave many, perhaps most RA patients in the lurch with respect to the promise of improved treatment in the near future. Even then, an epic drug once developed, may work for only a certain subset of RA patients, as in the case of anti-TNF therapy. This, then, requires multiple avenues of therapeutic pursuit to be performed in parallel, to get a sufficient number of irons in the fire for each given patient.
The Beyond Pathetic: It’s difficult to imagine that so little can be done to improve the lot of millions of patients experiencing a chronic, disabling disease such as RA in what is now the second decade of the 21st century. In my estimation, while genetic progress has been substantial, and will likely be of significant help in establishing new treatments, in the absence of significant unforeseen events, effective treatments will be unavailable for some time to come.
Disclaimer: I am not a physician, nor have received training in rheumatology. Not meant to be medical advice!
Note: This is the third installment of a guest series by Professor Bob West. Please read part 1 for an introduction to this topic.



Fascinating and very, very informative! I have no doubt he has found a key if not many keys here to use…now we just need to figure out how to use these keys to resolve this disease. We are fortunate to have him on our team!
I am reading these installments and wondering. It seems that we who struggle with this disease hold answers to only half of the question. We are genetically disposed to it – TRUE. But something happened to us that triggered the onset. It is fascinating for me to ponder what that was. I was not in the midst of some major stressful time that I can recall and I had not been ill. (Most likely many odd triggers exist.) But WHAT keeps these myriad triggers from starting up RA in others who are genetically disposed? I think that a wealth of information must exist in our female, 40+ family members who are well and have not yet had any reason to discover any propensity for RA lurking in their genes. I think that would be fascinating research. Find these “lucky” folks and let’s learn from them! Are none of them “type A” personalities who stress out about stuff? Are they super-health nuts who rarely eat fast food? What might they have in common to have dodged this bullet? Some other gene perhaps? Hmmmm. I just keep wondering who might embark on such a research journey? And where would they get the money to do it?
Suzy,
Some of the complexity behind the questions you pose I intend to address in two upcoming posts. These additional details may help you better understand the nature of the problem. By and large, however, the up-front answer to your hypothetical questions is that environmental determinants for RA, like many late-onset chronic diseases, remain a big black box. If I had to guess, I would say that the possibilities you raised are not among the major factors involved. But that’s just a guess.
23andMe v3 does provide data for:
rs6822844
rs660895
rs13192841
rs10499194
rs6920220
My watch list also contains:
rs2837960
rs6910071
rs2395175
rs3817964
Great series of articles that you have done here!
Thanks for that! Nice to see readers actually contributing real data to this discussion. I’m a firm believer in crowdsourcing improved healthcare.